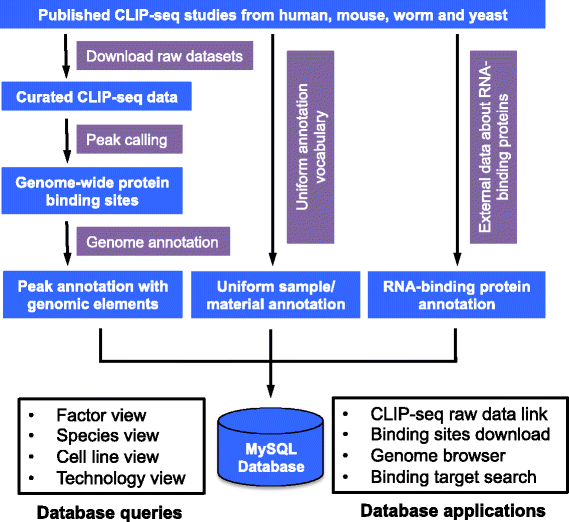
starBase v2.0: decoding Interaction Networks of lncRNAs, miRNAs, ceRNAs, RNA-Binding Proteins and mRNAs from large-scale tumor samples and CLIP-Seq (HITS-CLIP, PAR-CLIP, iCLIP, CLASH) data
Blockchain Network Line Icon. Nodes Connected Into Chain. Distributed Cloud Database For Secure Internet Transactions, Crypto Currency, Transfer Virtual Money Via Internet. Simple Minimal Pictogram. Vector Royalty Free Cliparts, Vectors, And Stock
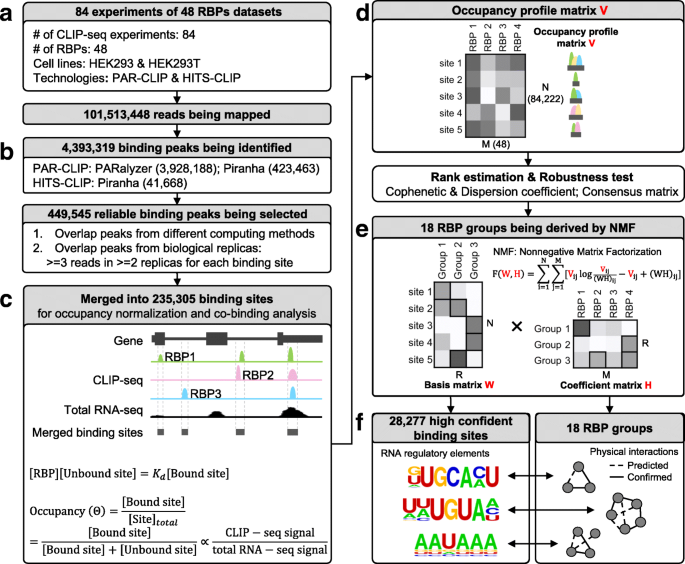
Identification of high-confidence RNA regulatory elements by combinatorial classification of RNA–protein binding sites | Genome Biology | Full Text

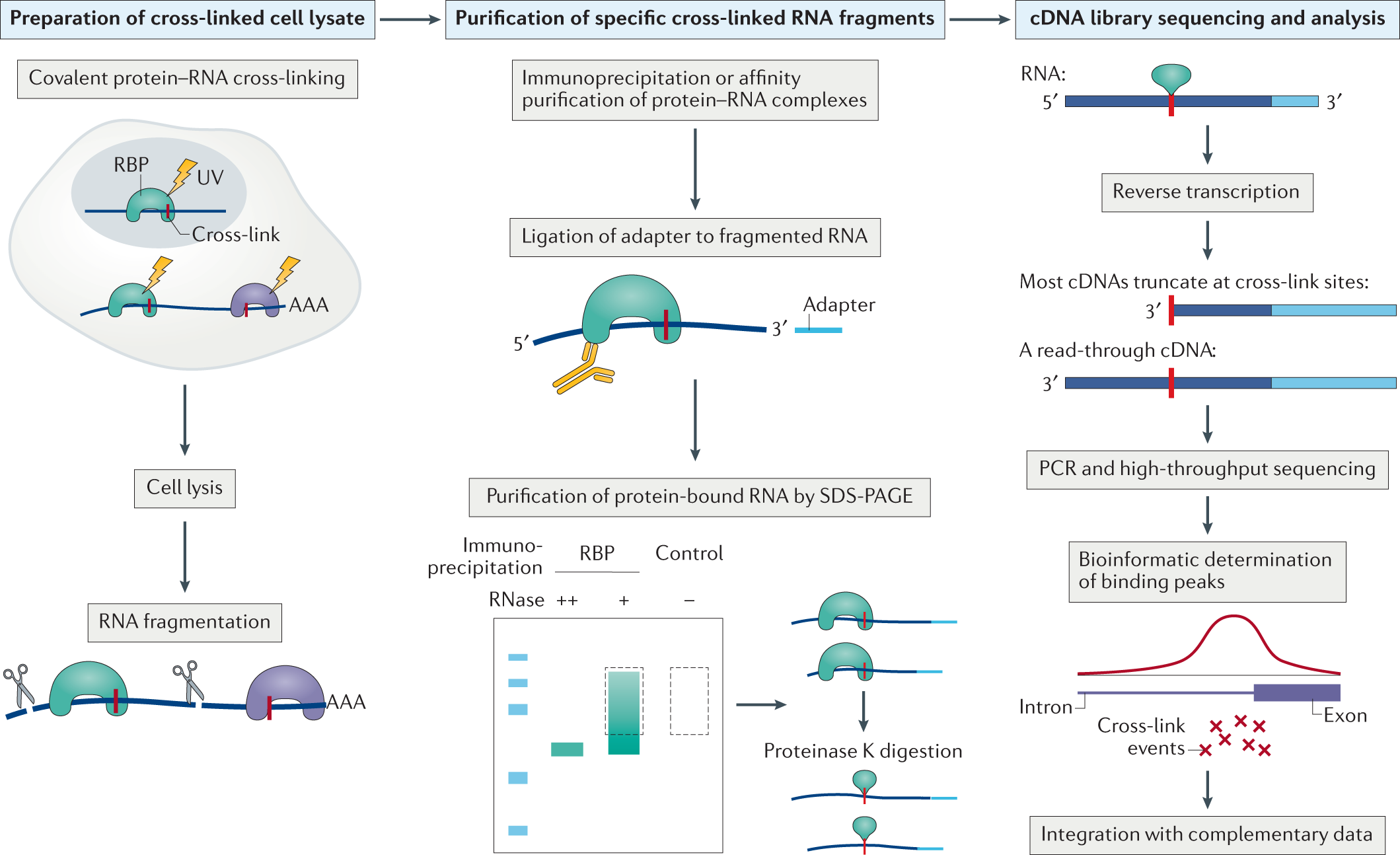
Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP: Cell

RNA-binding protein hnRNPLL regulates mRNA splicing and stability during B-cell to plasma-cell differentiation | PNAS

Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP: Cell

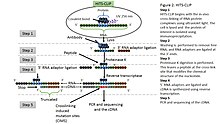
Outline of HITS-CLIP, PAR-CLIP and several variants, iCLIP, iCLAP and... | Download Scientific Diagram
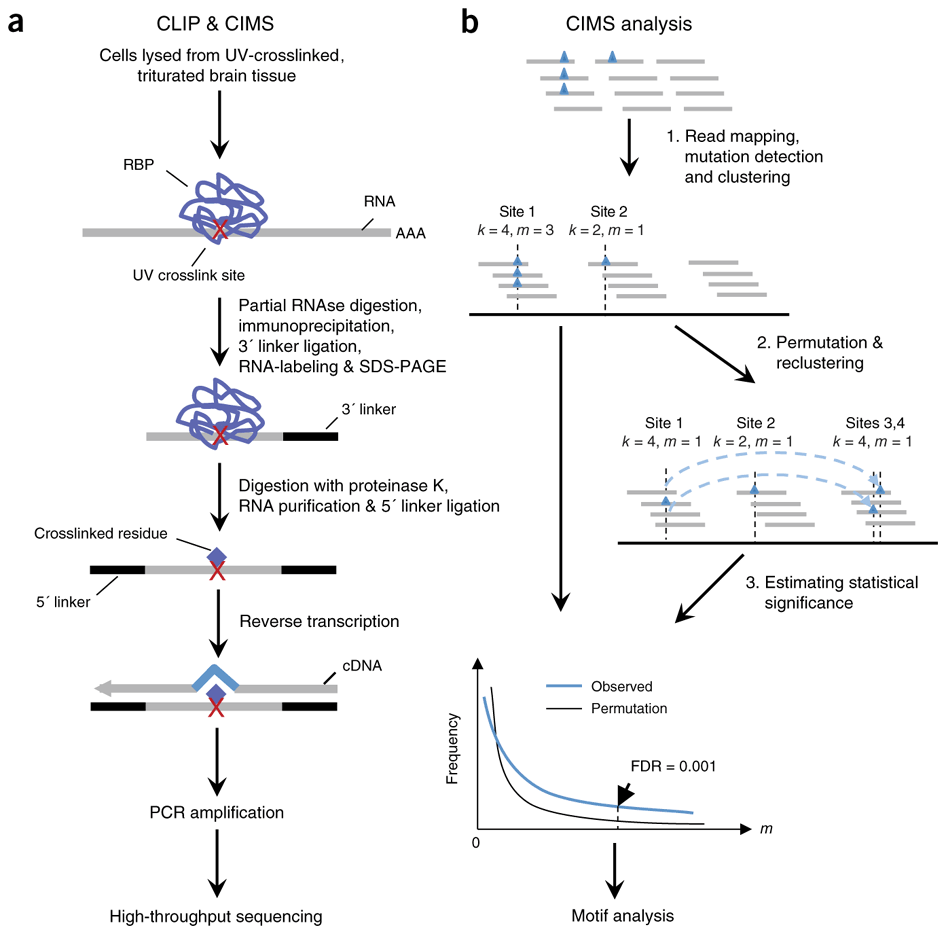
Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data | Nature Biotechnology
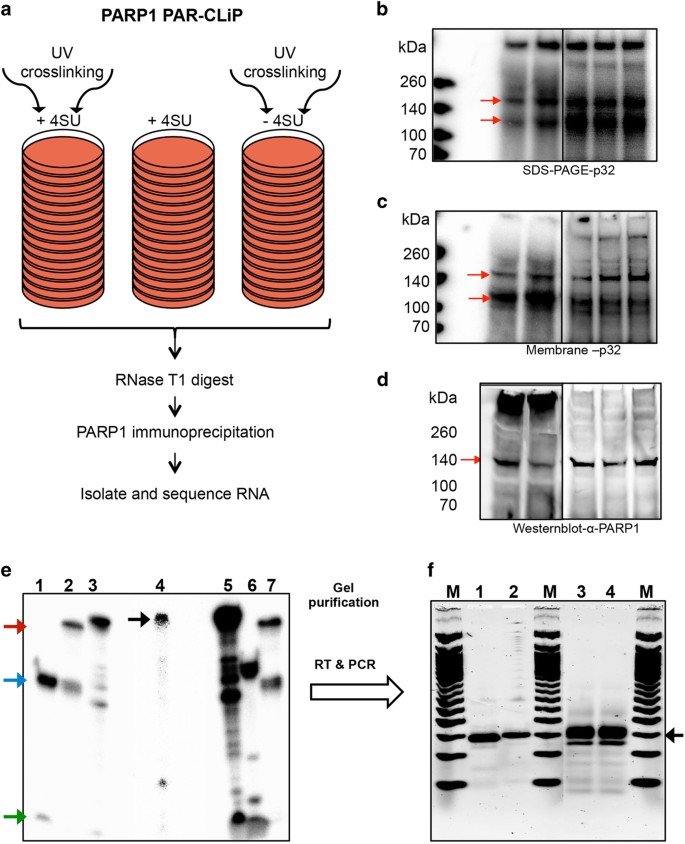
Transcriptome-wide identification of the RNA-binding landscape of the chromatin-associated protein PARP1 reveals functions in RNA biogenesis | Cell Discovery

Defining the RNA interactome by total RNA‐associated protein purification | Molecular Systems Biology

Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP - ScienceDirect


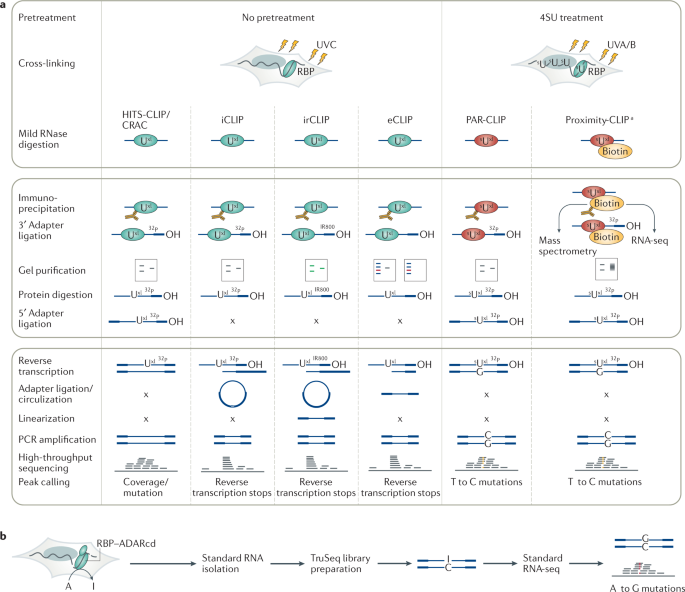
![The PARA-suite: PAR-CLIP specific sequence read simulation and processing [PeerJ] The PARA-suite: PAR-CLIP specific sequence read simulation and processing [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2016/2619/1/fig-1-full.png)